Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Wavelet-based Filter Bank Approach for Image Compression and Enhancement in Alzheimer\'s Disease Diagnosis Using Medical Imaging
Authors: Bhumika Mathur, Pankaj Shukla, Dr Mithlesh Kumar
DOI Link: https://doi.org/10.22214/ijraset.2024.63808
Certificate: View Certificate
Abstract
: This research presents a wavelet-based filter bank approach for image compression and enhancement in the context of Alzheimer\'s disease diagnosis using medical imaging. The proposed system leverages the PyWavelets and OpenCV libraries in Python to implement Discrete Wavelet Transform (DWT) and Inverse Discrete Wavelet Transform (IDWT) algorithms. The primary objective is to develop an efficient image compression technique while preserving visual quality and enhancing relevant features for improved diagnostic accuracy. The system utilizes filter banks to process wavelet coefficients, enabling selective enhancement of patterns or biomarkers associated with Alzheimer\'s disease. Results show that the decompressed images closely resemble the originals, with low Mean Squared Error (MSE) and high Peak Signal-to-Noise Ratio (PSNR) values. Visual inspection and quantitative metrics confirm the successful preservation of image quality and structural details. The wavelet coefficients visualization highlights the captured significant features crucial for reconstruction
Introduction
I. INTRODUCTION
Multimedia applications, scientific research, medical diagnostics, and other domains all heavily rely on image processing [1-7]. The significance of image processing in the field of medical imaging cannot be emphasized. Filtering, augmentation, segmentation, compression, and other techniques are essential for obtaining relevant information from medical images so that precise diagnosis and efficient treatment planning are possible. The analysis of imaging data from modalities such as positron emission tomography (PET), magnetic resonance imaging (MRI), computed tomography (CT) scans, and X-rays is one of the main uses of image processing in the medical profession [8-12]. Large volumes of data are produced by these imaging techniques, and image processing algorithms are necessary to improve the quality, lower noise, and extract pertinent characteristics from the images. Image processing can help radiologists and doctors identify anomalies, track the course of a disease, and decide on the best course of action by enhancing image clarity and emphasizing particular regions of interest. Also, image processing is essential for compressing medical images, which is necessary for effectively storing and sending huge imaging datasets. Both lossless and lossy compression methods, which are frequently based on wavelet modifications or other sophisticated algorithms, allow for large file size reductions without sacrificing the photos' diagnostic integrity. This allows for the easy sharing of medical pictures across healthcare providers and organizations, saving on storage space while promoting cooperative diagnosis and treatment planning.
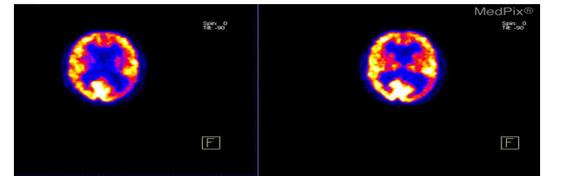
Beyond its use in medicine, image processing is essential to many scientific fields, including microscopy, astronomy, and remote sensing. Image processing techniques are utilized in these domains to improve the quality of acquired images, eliminate artifacts, and extract significant information from intricate data sets. For example, image processing techniques can be applied to astronomical imaging to enhance the visibility of celestial bodies, eliminate atmospheric distortions, and examine the characteristics of stars and galaxies [13-16]. Further, image processing is necessary for multimedia applications like computer vision, augmented reality, and picture and video editing. Features like object detection, picture recognition, and image enhancement are made possible by advanced algorithms and are essential to many contemporary applications and technologies.
In the field of medical imaging, filter banks have various potential uses, especially for the detection and tracking of neurodegenerative illnesses like Alzheimer's disease (AD). Feature extraction and image enhancement are two of the main uses. Filter banks can amplify modest anatomical or functional alterations in the brain linked to AD by selectively processing wavelet coefficients in particular frequency subbands. By doing this, biomarkers or patterns that might not be immediately visible in the original medical imaging can be better seen and detected. For example, filter banks can be made to emphasize the hallmark symptoms of AD, such as atrophy in the hippocampus or temporal lobe. Data compression is yet another essential use.
Medical imaging data can be quite vast and resource-intensive to store and transmit, especially from modalities like magnetic resonance imaging (MRI) and Positron Emission Tomography (PET). When used with wavelet-based compression technologies, filter banks can greatly reduce the size of these datasets without sacrificing any clinically meaningful information. This compression can help medical imaging data be efficiently stored, transmitted, and shared, facilitating quicker access to diagnostic data and optimizing workflows in healthcare institutions. In addition, filter banks can be used to remove artifacts and reduce noise in medical images. Various forms of noise or interference present in the imaging data can be selectively suppressed or eliminated by filter banks through the isolation and processing of particular wavelet coefficients. This can raise the overall level of clarity and quality in the photos, which will help doctors identify and interpret disorders like AD more correctly. Furthermore, filter banks can be customized for certain clinical uses or imaging modalities. For example, filter banks can be created to highlight or isolate regions or patterns of interest in functional imaging techniques like PET, which measure the brain's metabolic activity. This could potentially improve the diagnostic accuracy for conditions like AD, where metabolic changes in particular brain regions.
The main difficulty is in efficiently interpreting and extrapolating pertinent information from medical imaging data, especially when considering the minute structural and functional alterations linked to AD. The precise patterns or biomarkers suggestive of AD may not be well detected and enhanced by current image processing and analysis techniques, which could result in a delayed or incorrect diagnosis. Innovative image processing methods that can efficiently compress medical imaging data while maintaining critical diagnostic information and improving pertinent features are required to meet this challenge. Wavelet-based filter banks offer a possible way to accomplish these objectives. Filter banks have the ability to increase the visibility of AD-related patterns or biomarkers in medical images while simultaneously improving image compression efficiency by utilizing the multi-resolution capabilities of wavelet transforms and selectively manipulating wavelet coefficients. Nevertheless, the efficiency of various filter bank implementations in this particular setting has yet to be investigated. The effectiveness of different filter bank designs in terms of compression efficiency, maintaining picture quality, and highlighting AD-specific features in medical imaging data needs to be assessed and compared.
Thus, the goal of this research is to create an image compression system that is especially designed for medical imaging data linked to the diagnosis of Alzheimer's disease, using a wavelet-based filter bank technique. The Discrete Wavelet Transform (DWT) and Inverse Discrete Wavelet Transform (IDWT) algorithms will be implemented by the system using PyWavelets and OpenCV modules, allowing for effective compression without sacrificing visual quality. In order to improve the diagnostic accuracy of medical imaging techniques for Alzheimer's disease, this research intends to provide insights and prospective solutions by investigating the efficiency of various filter banks and their effects on compression efficiency and image quality.
II. EXPERIMENTAL PROCEDURE
The objective of this experiment was to implement and evaluate a wavelet-based image compression and decompression system using Python libraries, specifically OpenCV and PyWavelets, on PET images shown in Figure 1. The initial step in setting up the required environment was installing the necessary Python libraries, which include Matplotlib and NumPy for numerical operations and visualization, respectively, and OpenCV for image handling and PyWavelets for wavelet transformations. The application was then loaded with the sliced images. To make things simple, OpenCV was used to read the photos in grayscale mode, which works well for intensity-based analysis. The grayscale image was first compressed by applying the Discrete Wavelet Transform (DWT), which divided the image into its wavelet coefficients. The image is represented by these coefficients at various resolution settings. The DWT was carried out using the PyWavelets module, and the Haar wavelet was chosen for this particular experiment.
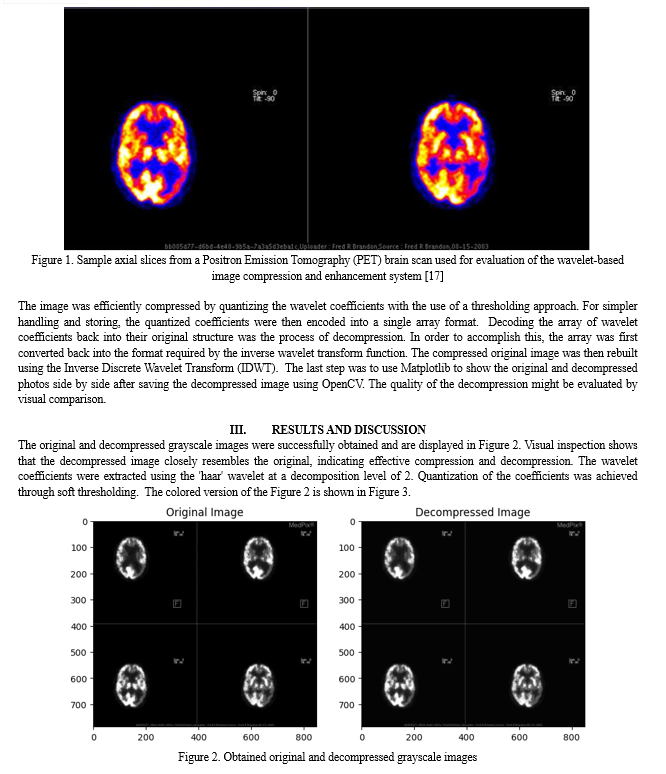
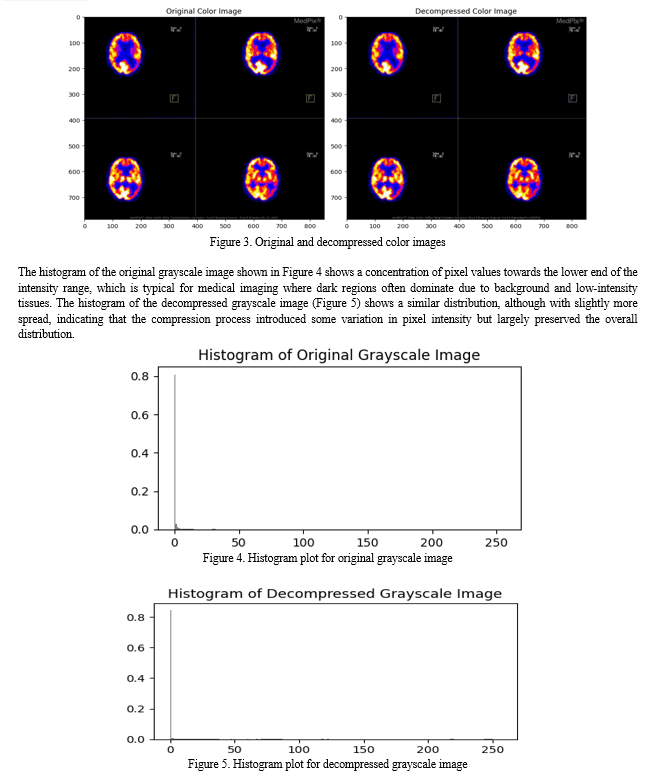
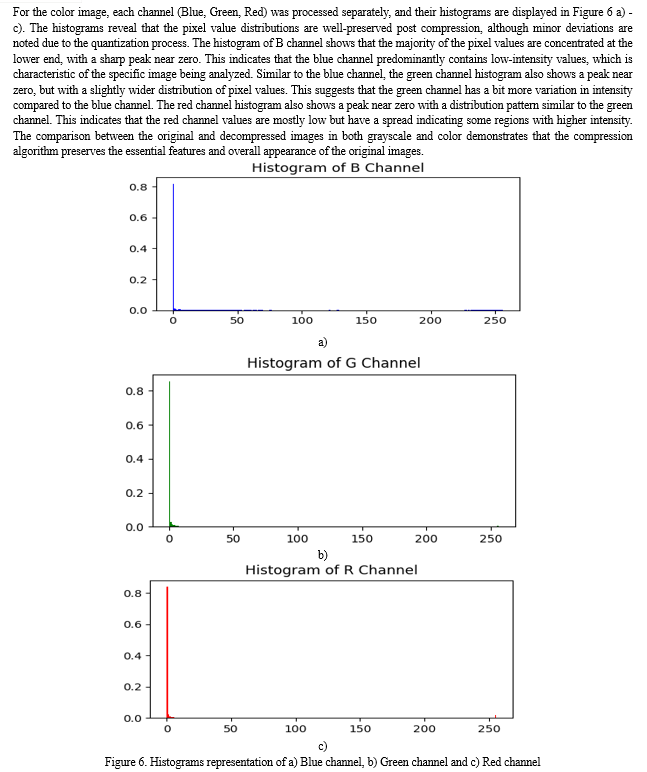

Conclusion
The presented research successfully implemented and evaluated a wavelet-based filter bank approach for image compression and enhancement in the context of Alzheimer\'s disease diagnosis using medical imaging. The proposed system leveraged the PyWavelets and OpenCV libraries in Python to implement the Discrete Wavelet Transform (DWT) and Inverse Discrete Wavelet Transform (IDWT) algorithms. Visual inspection and quantitative metrics confirmed the preservation of image quality and structural details. The wavelet coefficients visualization highlighted the captured significant features crucial for reconstruction, suggesting the potential for selective enhancement of Alzheimer\'s disease-related patterns or biomarkers.
References
[1] Chauhan, R.B., Shah, T.V., Shah, D.H., Gohil, T.J., Oza, A.D., Jajal, B. and Saxena, K.K., 2023. An overview of image processing for dental diagnosis. Innovation and Emerging Technologies, 10, p.2330001. [2] Boopathi, S., Pandey, B.K. and Pandey, D., 2023. Advances in artificial intelligence for image processing: techniques, applications, and optimization. In Handbook of research on thrust technologies’ effect on image processing (pp. 73-95). IGI Global. [3] Dhar, T., Dey, N., Borra, S. and Sherratt, R.S., 2023. Challenges of deep learning in medical image analysis—Improving explainability and trust. IEEE Transactions on Technology and Society, 4(1), pp.68-75. [4] Huang, Z., Bianchi, F., Yuksekgonul, M., Montine, T.J. and Zou, J., 2023. A visual–language foundation model for pathology image analysis using medical twitter. Nature medicine, 29(9), pp.2307-2316. [5] Burger, W. and Burge, M.J., 2022. Digital image processing: An algorithmic introduction. Springer Nature. [6] Van der Velden, B.H., Kuijf, H.J., Gilhuijs, K.G. and Viergever, M.A., 2022. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Medical Image Analysis, 79, p.102470. [7] Tov, O., Alaluf, Y., Nitzan, Y., Patashnik, O. and Cohen-Or, D., 2021. Designing an encoder for stylegan image manipulation. ACM Transactions on Graphics (TOG), 40(4), pp.1-14. [8] Manoharan, S., 2020. Early diagnosis of lung cancer with probability of malignancy calculation and automatic segmentation of lung CT scan images. Journal of Innovative Image Processing (JIIP), 2(04), pp.175-186. [9] Zunair, H., Rahman, A., Mohammed, N. and Cohen, J.P., 2020. Uniformizing techniques to process CT scans with 3D CNNs for tuberculosis prediction. In Predictive Intelligence in Medicine: Third International Workshop, PRIME 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 8, 2020, Proceedings 3 (pp. 156-168). Springer International Publishing. [10] Oulefki, A., Agaian, S., Trongtirakul, T. and Laouar, A.K., 2021. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern recognition, 114, p.107747. [11] Serena Low, W.C., Chuah, J.H., Tee, C.A.T., Anis, S., Shoaib, M.A., Faisal, A., Khalil, A. and Lai, K.W., 2021. An overview of deep learning techniques on chest X-ray and CT scan identification of COVID-19. Computational and Mathematical Methods in Medicine, 2021, pp.1-17. [12] Wang, M., Yang, X. and Wang, W., 2022. Establishing a 3D aggregates database from X-ray CT scans of bulk concrete. Construction and Building Materials, 315, p.125740. [13] Uzeirbegovic, E., Geach, J.E. and Kaviraj, S., 2020. Eigengalaxies: describing galaxy morphology using principal components in image space. Monthly Notices of the Royal Astronomical Society, 498(3), pp.4021-4032. [14] Farias, H., Ortiz, D., Damke, G., Arancibia, M.J. and Solar, M., 2020. Mask galaxy: Morphological segmentation of galaxies. Astronomy and Computing, 33, p.100420. [15] Nightingale, J., Amvrosiadis, A., Hayes, R.G., He, Q., Etherington, A., Cao, X., Cole, S., Frawley, J., Frenk, C.S., Lange, S. and Li, R., 2023. PyAutoGalaxy: Open-Source Multiwavelength Galaxy Structure & Morphology. Journal of Open Source Software, 8(81). [16] Zaritsky, D., Donnerstein, R., Karunakaran, A., Barbosa, C.E., Dey, A., Kadowaki, J., Spekkens, K. and Zhang, H., 2022. Systematically Measuring Ultra-diffuse Galaxies (SMUDGes). III. The Southern SMUDGes Catalog. The Astrophysical Journal Supplement Series, 261(2), p.11. [17] https://medpix.nlm.nih.gov/topic?id=0696442f-28c1-4bfb-83a0-8e7543b7f1af
Copyright
Copyright © 2024 Bhumika Mathur, Pankaj Shukla, Dr Mithlesh Kumar. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET63808
Publish Date : 2024-07-30
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

